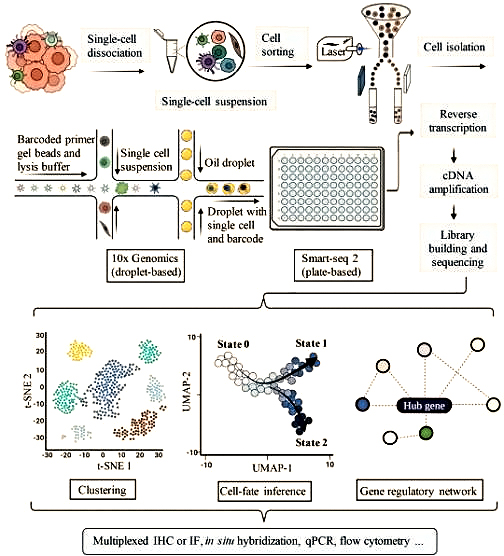
Traditional second-generation sequencing (NGS) examines the genome of a population of cells, such as cell culture, tissues, organs, or the entire organism. Its output is the 'average genome' of the cell population, while single-cell sequencing measures the genome of individual cells in the cell population. Nowadays, traditional methods are referred to as batch sequencing to distinguish them from single-cell technology. At present, single-cell sequencing technology can be used to measure the genome (scDNA seq), DNA methylation group, or transcriptome (scRNA seq) of each cell in a population. These techniques have been used to identify new mutations in cancer cells, explore progressive epigenetic changes that occur during embryonic development, and evaluate how a seemingly homogeneous population of cells expresses specific genes. In single-cell analysis methods, scRNA seq comprehensively measures the transcriptional expression of large volume cells. And quantitatively analyze all transcripts expressed in individual cells, providing an unbiased strategy for identifying and describing different cell populations through scRNA sequence analysis of organ rejection reactions. After tissue biopsy, cells are isolated and can be used for droplet or plate based sequencing methods. After batch effect correction, standardization, data imputation, and dimension adjustment, specific cell subtypes can be determined. Heterogeneity analysis and tolerance prediction are used to determine target gene and molecular interactions, which are the basis for gene therapy methods and long-term transplant acceptance.